Gene Regulation and Gene Expression: Differences
Regulation of gene expression includes many mechanisms that are used by the cells to increase or decrease the production of specific gene products. The different programs of gene expression in biology, e.g., to initiate developmental pathways, and respond to environmental stimuli, or for adaptation. Any of those steps of gene expression are modulated, from transcriptional regulation, to RNA processing, and to post-translational modification of protein.
This Story also Contains
- What is Gene Regulation?
- Genes
- Gene Expression
- Regulation of Gene Expression
- Prokaryotic and Eukaryotic Transcription
- Applications of Gene Regulation and Expression
- Recommended video for "Gene Regulation"
- MCQs on Gene Expression and Gene Regulation

Gene regulation is important for viruses, prokaryotes, and eukaryotes as it increases the versatility of an organism by allowing the cell to synthesise proteins when needed. The first discovery of the gene regulation system is widely considered to be the identification of the lac operon in 1961. It was discovered by Francis Jacob and Jacques Monod, in which some enzymes involved in glucose metabolism are expressed by E.coli. Gene expression and gene regulation are topics of the chapter Molecular Basis of Inheritance. It is an important chapter in the Biology subject.
What is Gene Regulation?
Gene regulation refers to the set of steps that control the rate and fashion in which genes are expressed. Gene expression is the process by which information from an individual gene is used to synthesise a functional gene product, often proteins. These processes are vitally essential for cellular functions in determining cell behaviour and global organism development. Appropriate gene regulation confers the ability to express genes at the right time, location, and level of activity to uphold homeostasis and in response to environmental change.
This article explicates in detail the genes and their expressions and regulations. It, therefore, considers the mechanisms of expression of genes and the comparison of the process of transcription in prokaryotes and eukaryotes, applications and implications of gene regulation in medicine, biotechnology, and even evolution.
Genes
Genes are segments of deoxyribonucleic acid (DNA) encoding information for the synthesis of proteins or sometimes functional RNA molecules. Genes represent the elementary units of heredity and, hence, master the determination of determining characteristics of organisms. The two main types of regions that make up genes are coding and regulatory regions.
Genes contain a sequence of exons, introns, and regulatory elements—in particular, promoters, enhancers, and silencers. The exons are the sequences that code for proteins. Introns are non-coding and are spliced out during the process of RNA processing. Promoters are DNA sequences that define the start site for transcription, whereas enhancers increase the probability of transcription and silencers decrease it.
Rather, the human genome encodes some 20,000 to 25,000 protein-coding genes. For that matter, the mechanisms of gene expression and regulation show such a high degree of complexity that the diversity of produced proteins and functional outcomes is immensely wider than this mere count would suggest. In consequence, a fine-tuned network ensures the normal functioning of cells and the development of organisms by the interplay between genes, regulatory elements, and environmental factors.
Gene Expression
Gene expression is defined as the process by which genetic information contained in an individual's genome is used for protein synthesis or the formation of functional RNA. This mostly occurs in two main stages: transcription and translation. Transcription is the process of converting a gene's DNA sequence into a messenger RNA transcript. Translation is the decoding of the mRNA by the ribosome to synthesise a linear chain of amino acids into protein.
Transcription is located in the eukaryotic nucleus and the cytoplasm of prokaryotes. This process gets started as RNA polymerase binds to a gene's promoter region, which eventually causes the DNA helix to separate, followed by the synthesis of an RNA strand that is complementary in base pairing under the template strand. This primary transcript, in eukaryotes, is processed by the addition of a 5' cap, polyadenylation at the 3' end, and splicing to remove the introns and join the exons.
Translation occurs in the cytoplasm: The reading of mRNA sequences is done by ribosomes in translation, further assembling an amino acid chain according to such sequence. tRNA molecules bring the appropriate amino acids to the ribosome according to the codons of a sequence in mRNA. The formed polypeptide folds into a conformation, making a functional protein, which can carry forward cellular functions native to it.
Regulation of Gene Expression
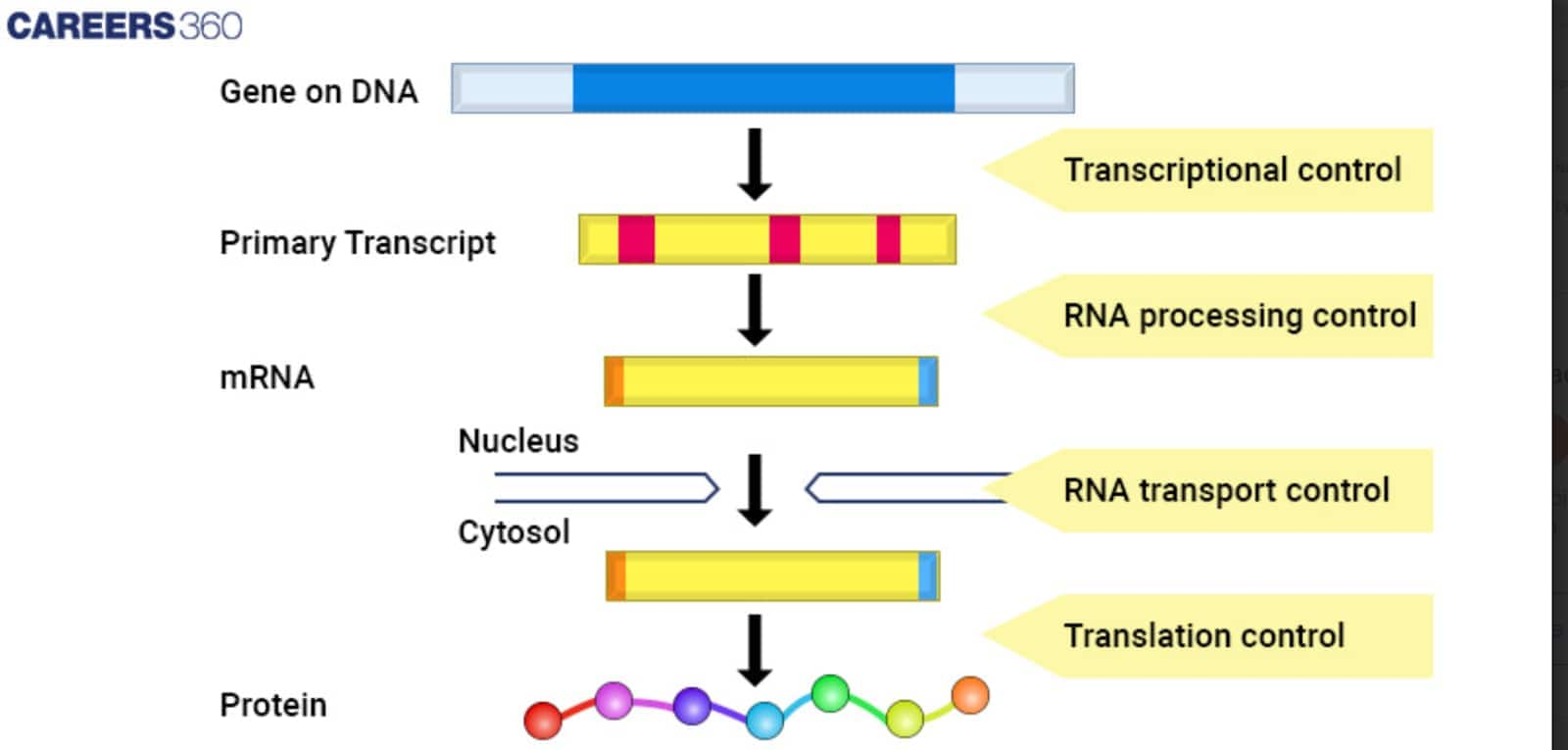
Gene expression is in an extremely regulated manner at various levels: chromatin, transcriptional control, post-transcriptional modifications of RNA, the process of translation, and post-translational modifications of proteins. These are the mechanisms controlling the processes to make sure the genes are expressed in the right cells at the right times and in proper amounts.
Chromatin Structure And Epigenetics
The structure of the chromatin, made up of DNA bound to histone proteins, plays a very important role in gene expression. Changes in the structure of chromatin or accessibility to genes and the machinery involved in transcription can occur through histone modifications, such as acetylation and methylation of the lysine tails. Another form of epigenetic modification is the addition of a methyl group to the DNA, usually leading to the silencing of gene expression by occluding the binding of transcription factors to DNA.
Transcriptional Control
Transcription factors are proteins that attach themselves to certain DNA sequences, thus regulating the genes that would then be involved with transcription. They could either act as activators, increasing the rate of transcription, or as repressors, decreasing the rate of transcription. The rate of gene transcription is established by the binding of transcription factors in the promoter, enhancer, and silencer elements.
Post-Transcriptional Modifications
After transcription, the initial RNA transcript is modified in several ways before it can be translated as mature mRNA. Splicing excises the introns and juxtaposes the exons, and alternative splicing can generate multiple mRNA isotypes or variants from one gene, thus representing an additional level of potential protein diversity. The 5′cap site and poly-A tail are both added to the mRNA to help stabilise it and assist in its export from the nucleus.
Translational Control
Regulation takes place during the initiation, elongation, and termination phases of translation. Translation initiation factors, ribosomal subunits, and tRNA molecules are involved in ensuring that the initiation phase of translation occurs effectively and with correct precision. Regulatory proteins interact with mRNA to control translation into protein.
Post-translational Modifications
Post-translation, proteins undergo various further modifications including phosphorylation, glycosylation, and ubiquitination, which cause changes in the activity, stability, subcellular localisation, and function. This implies that each event confers a specificity that tells the cell to perform certain functions.
Diagram: Gene regulation
The diagram given below indicates the process involved in Gene Expression and Regulation:
Prokaryotic and Eukaryotic Transcription
Transcription, the process by which a single-stranded RNA is synthesised given a double-stranded DNA molecule, differs significantly in many ways between prokaryotes and eukaryotes. These differences reflect the cellular components that vary between the two domains of life and the different mechanisms of regulation characteristic of them.
Feature | Prokaryotic Transcription | Eukaryotic Transcription |
Location | Cytoplasm | Nucleus |
RNA Polymerases | Single RNA polymerase | Multiple RNA polymerases (I, II, III) |
Initiation | Simple initiation process | Complex initiation with multiple transcription factors |
Promoters | Simple promoter regions | Complex promoters with TATA boxes and other elements |
mRNA Processing | No processing (mRNA is used directly) | Extensive processing (capping, polyadenylation, splicing) |
Transcription and Translation | Coupled (occur simultaneously) | Uncoupled (transcription in nucleus, translation in cytoplasm) |
Applications of Gene Regulation and Expression
Gene regulation and expression form the basis of the intricate functioning of biological systems, influencing everything from basic cellular processes to the evolution of complex traits and diseases. Here's an exploration of their applications across different fields:
Medical Applications
Gene regulation is a very important consideration in the understanding and subsequent treatment of diseases like cancer and a host of other genetic disorders. Aberrant gene expression can foster out-of-control growth of cells, leading to cancer. In understanding these regulatory mechanisms, scientists can go on to synthesise gene therapy in a bid to correct defective genes or, using CRISPR technology, edit a gene to treat a genetic disorder.
Gene regulation is central in genetic engineering, in the making of GMOs, and in synthetic biology to engineer traits of interest in crops. It is also required for the production of proteins for human and animal pharmaceutical applications, and in engineered industrial microorganisms.
Evolutionary Implications
Gene regulation can be a driving force throughout evolution and development. Changes in regulatory elements can lead to variations in expression at the level of genes, contributing to the diversity of life forms. Understanding these regulatory mechanisms finally gives pieces of information on how complex traits and organisms have evolved.
Recommended video for "Gene Regulation"
MCQs on Gene Expression and Gene Regulation
Q1. In gene regulation, open reading frame (ORF) implies
Option 1: intervening nucleotide sequence in between two genes.
Option 2: a series of triplet codons not interrupted by a stop codon.
Option 3: a series of triplet codons that begins with a start codon and ends with a stop codon.
Option 4: the exonic sequence of a gene that corresponds to the 5'UTR of the mRNA and thus does not code for the protein
Correct answer: 2) a series of triplet codons not interrupted by a stop codon.
Explanation:
In gene regulation, an open reading frame (ORF) refers to a DNA sequence that has the potential to be translated into a protein. It is a continuous series of triplet codons that are not interrupted by a stop codon.
The start codon (usually AUG, which codes for methionine) marks the beginning of an ORF. From the start codon, the ribosome reads the mRNA sequence in a 5' to 3' direction, synthesising a protein until it reaches a stop codon (UAA, UAG, or UGA), which signals the end of translation.
An ORF is considered to be a potential protein-coding sequence if it contains a start codon followed by a series of triplet codons without any intervening stop codons. This uninterrupted sequence suggests that the reading frame has the potential to be translated into a functional protein.
Hence, the correct answer is option 2) a series of triplet codons not interrupted by a stop codon.
Q2. Control of gene expression in prokaryotes takes place at the level of:
Option 1: DNA replication
Option 2: Transcription
Option 3: Translation
Option 4: None of the above
Correct answer: 2) Transcription.
Explanation:
The control of gene expression in prokaryotes is mainly effectuated at the transcription level, which involves the regulation of RNA synthesis from DNA. This is predominantly achieved by modulating the initiation of transcription through the use of regulatory sequences such as promoters, operators, and repressors.
Operons are characteristic of prokaryotic gene regulation. An operon is a group of functionally linked genes that are co-transcribed. Its expression is controlled by a regulatory gene, which can produce either a repressor or an activator protein.
A prime illustration is the lac operon in E. coli, which oversees lactose metabolism. In the absence of lactose, the repressor protein obstructs transcription. However, when lactose is present, the repressor is inactivated, thus facilitating transcription.
Additionally, transcription factors such as activators and repressors play a pivotal role. These molecules can either enhance or hinder RNA polymerase binding to DNA, thereby controlling transcription initiation.
Hence, the correct answer is option 2) Transcription.
Q3. Factors regulating genes expression are,
Option 1: Metabolic Physiology
Option 2: Environmental Condition
Option 3: Both of these
Option 4: None of these
Correct answer: 3) Both of these
Explanation:
Metabolic, physiological, or environmental conditions influencing the expression of genes are factors that regulate gene expression. Regulation in this regard is of utmost importance for the development and differentiation of the embryo into an adult organism since the process involves the coordinated expression of sets of genes. This ensures that the appropriate sets of genes get activated or repressed at the right time and in the appropriate context, thereby allowing for proper growth and function.
Hence, the correct answer is Option 3) Both of these
Also Read-
Frequently Asked Questions (FAQs)
Gene expression, i.e., transcription of DNA into mRNA and translation of mRNA into protein, gives rise to the distinct operations of cell type.
Gene regulation mechanisms encompass different modulations in chromatin structure, transcriptional control, post-transcriptional modifications, translational control, and post-translational modifications.
Prokaryotic gene regulation is simple, which happens in the cytop[lasm without processing of the mRNA, whereas eukaryotic regulation is a bit complex for many RNA polymerases, transcription factors, and enormous RNAm processing.
Gene therapy, application of CRISPR for gene editing, generating GMOs, synthetic biology in the production of therapeutic proteins, and microorganism engineering.
It functions by controlling when, where, and how much gene expression is required for normal cellular functioning and development.