Restriction Enzyme: Definition, Types, Applications, Examples, Diagram
Restriction enzymes are enzymes that cut DNA at specific sequences, known as recognition sites. It was discovered in bacteria. These enzymes play a vital role in defending against bacteriophages by cutting foreign DNA. In biotechnology, restriction enzymes are essential tools that allow the cutting and manipulation of DNA, helping in genetic experiments and advances in medicine and agriculture. These enzymes also help in understanding bacterial genetics, where their natural function was first observed.
This Story also Contains
- What are Restriction Enzymes?
- Types of Restriction Enzymes
- Mechanism of Action
- Specificity and Recognition Sites
- Applications in Molecular Biology
- Laboratory Techniques Involving Restriction Enzymes
- Advances and Innovations
- Practical Considerations in Using Restriction Enzymes
- MCQs on Restriction Enzyme
- Frequently Asked Questions (FAQs)
- Recommended Video on Restriction Endonuclease

In genetic engineering, restriction enzymes are used to create recombinant DNA by inserting desired genes into cloning vectors, such as plasmids. This is a critical step in the study and application of gene regulation and gene expression. Today, restriction enzymes are indispensable in labs for gene editing, DNA fingerprinting, and the development of genetically modified organisms (GMOs).
What are Restriction Enzymes?
Restriction enzymes or restriction endonucleases, are enzymes produced by bacteria that can cleave DNA molecules at or near specific base sequences. They are crucial in molecular biology because of their cutting of DNA at specific recognition sites. They were isolated in the 1960s and signalled more change in genetic engineering since they provided the exact method for manipulating DNA molecules. These enzymes are widely used in different molecular approaches, for instance in cloning, gene modification, and even DNA profiling.
Restriction enzymes are enzymes produced by bacteria and archaea that recognise and then cleave DNA. They also serve to cut the DNA molecule into fragments, where every fragment has an end or terminal boundary. Restriction enzymes have no doubt become one of the most crucial instruments in the context of molecular biology, especially regarding the manipulation and analysis of DNA. It lets researchers map the genomic structure, analyse the sequences of genes, and construct new DNA molecules or modified organisms.
The restriction enzyme is a protein produced by bacteria that cleaves DNA at specific locations. This location is known as the restriction site. They were first identified in the middle of the 1960s, basically through research on the mechanisms by which bacteria are against viruses. Werner Arber, Daniel Nathans, and Hamilton O. Smith shared the Nobel Prize for Physiology or Medicine for their discoveries concerning the restriction of bacterial viruses, which provided a new tool for analysing genetic material.
In general, restriction enzymes occur in bacteria and in some archaea, where they are part of the restriction-modification system. While restriction enzymes cut any foreign DNA that might invade the host cell, methyltransferases ‘protect’ host DNA from cutting itself by wrapping it around. This system has proved relevant in preventing viral infections and preventing bacteria from swapping genes.
Commonly Asked Questions
The term "restriction" comes from their natural function in bacteria, where they restrict the growth of viruses by cutting up foreign DNA. Bacteria use these enzymes as a defense mechanism against viral infections.
Types of Restriction Enzymes
Table: Comparison of Different Types of Restriction Enzymes or restriction endonucleases,
Feature | Type I Restriction Enzymes | Type II Restriction Enzymes | Type III Restriction Enzymes | Type IV Restriction Enzymes |
Recognition Sequence | Specific, bipartite sequences | Specific, palindromic sequences | Specific, short sequences | Modified DNA (e.g., methylated or glucosylated) |
Cleavage Site | Random, distant from the recognition site | Within or close to the recognition site | A short distance away from the recognition site | Modified DNA sites |
Cofactors Required | ATP, S-adenosylmethionine (SAM), Mg2+ | Mg2+ (sometimes other divalent cations) | ATP, Mg2+ | Mg2+ |
Subunit Composition | Complex, multi-subunit | Simple, usually a single subunit | Multi-subunit | Multi-subunit |
Cleavage Mechanism | DNA translocation and looping | Direct cleavage | DNA looping and cleavage | Specific for modified DNA |
Example Enzymes | EcoKI, EcoBI | EcoRI, HindIII, BamHI | EcoP15I, Hindi | McrBC, EcoKMcrA |
Applications | Limited due to non-specific cleavage | Widely used in molecular cloning, gene editing | Limited, specialized applications | Study of DNA modifications, epigenetics |
Resistant enzymes based on recognition sites, cleavage recognition sites, and mechanisms of action are distinguished.
Type I
Recognition Sites: Some restriction enzymes are of Type I that cause DNA cleavage at a random position away from the site of recognition but recognise a specific sequence.
Cleavage Pattern: It must be noted that they generate fragments of variable size because they cleave at non-specific sites.
Mechanism of Action: The newly discovered type I restriction enzymes are multi-subunit enzymes whose subunits include the recognition, cleavage, and modification subunits. They need ATP for the cleavage of DNA and possess the characteristics of both endonuclease and methyl transferase enzymes.
Type II
Recognition Sites: The Type II restriction enzymes act on DNA at or near the specific sequence at which the enzyme can recognize the DNA molecules.
Cleavage Pattern: They reproduce DNA segments and provide specialised terminal characteristics to the ends of the segments as blunt ends or sticky ends, according to the enzyme type.
Mechanism of Action: Type II restriction enzymes exist as homodimers or homotetramers and cleave DNA at specific sites, and no energy in the form of ATP is utilised. They are generally used in molecular biology for genetic engineering.
Type III
Recognition Sites: Type III restriction enzymes are specific in recognising DNA sequences, and there is DNA cleavage at a certain distance from the recognition site.
Cleavage Pattern: They produce DNA fragments with definite cut sites, like Type II enzymes.
Mechanism of Action: Type III restriction enzymes act as multicomponent molecules that consist of subunits that are designed for the recognition of the target DNA, the cutting of the DNA bonds, and the modification of DNA.
Type IV
Recognition Sites: Type IV restriction enzymes are those that cut DNA at specific base sequences on the DNA but do not use any ATP.
Cleavage Pattern: They cleave DNA non-specifically and produce DNA fragments with different lengths from those of the restriction endonucleases.
Mechanism of Action: Type IV restriction enzymes are endonucleases that make an incision on various DNA sequences and do not require ATP. They are considerably rare as compared to Type I, II, and III enzymes.
Commonly Asked Questions
Type II restriction enzymes are the most commonly used in biotechnology. They cut DNA at specific sites within or very close to their recognition sequences. Type I and Type III enzymes are more complex, requiring ATP and cutting DNA at variable distances from their recognition sites.
Isoschizomers are restriction enzymes from different bacteria that recognize and cut the same DNA sequence. Neoschizomers recognize the same sequence but cut at different positions.
Mechanism of Action
The mechanisms of action include the recognition of specific DNA sequences and cleavage patterns:
Recognition of Specific DNA Sequences
Restriction enzymes bind to a particular sequence on the DNA molecules and they are usually palindromic, This involves complementary base pairing between the enzyme and the DNA. The recognition sequence is different for all the enzymes and generally, it lies for four to eight nucleotide pairs.
Cleavage Patterns (Blunt Ends and Sticky Ends)
Once the particular sequence of the DNA has been identified by the restriction enzymes, they cut the DNA at specific sites separated by the particular base sequence, which causes either the formation of blunt ends or sticky ends. Cut ligation description Blunt ends: ends that are created when both of the DNA’s strands have been cleaved and a finish with a 90° angle is present, while sticky ends: are generated when the DNA is cleaved asymmetrically, leading to the presence of overhanging single strands of DNA. There are two types of cleavage patterns, depending on the enzymes to be used and the recognition sequence
Commonly Asked Questions
Bacteria use restriction enzymes to cut up foreign DNA, such as that from invading viruses. The bacteria's own DNA is protected by methylation, which prevents the restriction enzymes from cutting it.
Endonucleases, like restriction enzymes, cut DNA within the strand. Exonucleases, on the other hand, remove nucleotides from the ends of DNA strands. Restriction enzymes are a type of endonuclease.
Specificity and Recognition Sites
Explanation Of Recognition Sequences
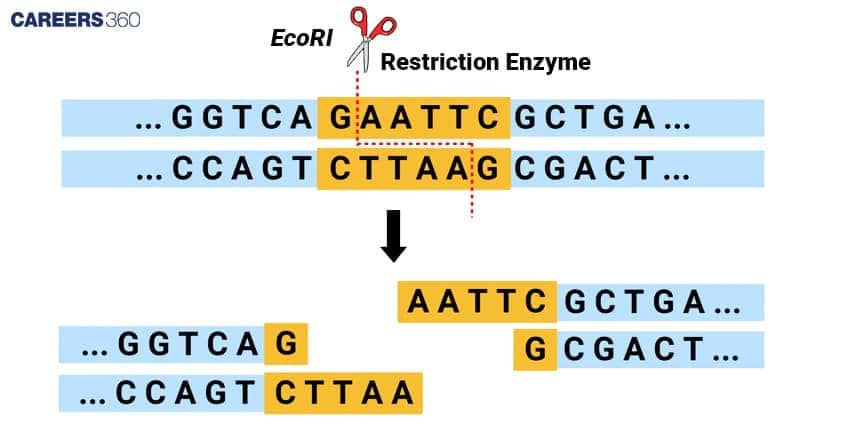
Recognition sequences are definable DNA sequences to which the restriction enzymes adhere and scission. These sequences are generally palindromic, as are the sequences on two complementary strands of DNA. For example, the recognition sequence for the restriction enzyme EcoRI is 5'-GAATTC-3', which reads the same on both strands: The reverse complement sequence is the ‘5’ to ‘3’ orientation, stated as ‘5’-GAATTC-‘3’.
Palindromic Sequences
Palindromes have a sequence; on one aspect, it is exactly the reverse of the other aspect of the other strand. This symmetry enables the restriction enzymes to efficiently bind to the DNA at precise sites to cleave the DNA and give predictable cleavage patterns.
Commonly Asked Questions
Restriction enzymes recognize specific DNA sequences, typically 4-8 base pairs long, called restriction sites. They bind to these sites and cleave the DNA strand at or near this recognition sequence.
A palindromic sequence in DNA reads the same forwards and backwards on complementary strands. Many restriction enzymes recognize these sequences. For example, the EcoRI recognition site GAATTC is palindromic.
The recognition site is the specific DNA sequence that a restriction enzyme binds to. The cleavage site is where the enzyme actually cuts the DNA, which may be within or near the recognition site, depending on the enzyme.
Frequent cutters are restriction enzymes that recognize short DNA sequences (usually 4 base pairs), cutting DNA more often. Rare cutters recognize longer sequences (6 or more base pairs), cutting DNA less frequently.
Some restriction enzymes are sensitive to DNA methylation, a key epigenetic modification. By comparing digestion patterns of methylation-sensitive and insensitive enzymes, scientists can study DNA methylation patterns.
Applications in Molecular Biology
Genetic engineering
Under this category, restriction enzymes are very vital in genetic engineering since they can be used to alter DNA sequences. They are employed to cut the DNA at a particular site of recognition to enable the addition, removal, or alteration of DNA segments. This ensures the creation of recombinant DNA molecules with desired traits for various applications.
Cloning
To clone genes, restriction enzymes are applied to the vector DNA, and the DNA insert has to be cut at particular sites (40). Because they have sticky ends, the vector and insert DNA fragments can recombine by base pairing, resulting in the formation of recombinant DNA molecules. These recombinant molecules are then taken into host cells for replication as well as expression of the inserted gene.
Recombinant DNA Technology
Recombinant DNA is the result of the formation of new DNA sequences from DNA fragments derived from different sources. In this process, restriction enzymes are crucial because they cut the DNA at predetermined sites and help in linking together new pieces of DNA with desirable properties.
DNA fingerprinting
In DNA fingerprinting, restriction enzymes can cut the genomic DNA randomly into smaller pieces of DNA fragments of different sizes. The obtained DNA fragments are run on a gel for electrophoresis, which generates specific band patterns that are referred to as fingerprints. This technique is used routinely, especially in the fields of forensics, DNA paternity testing, and DNA profiling.
CRISPR Technology
CRISPR also called cluster regularly Inter-spaced palindromic repetitive sequences is a remarkable modification that employs the capability of restriction enzymes for molecular sickleization. CRISPR-associated (Cas) proteins cut target DNA sequences that base pair with complementary RNA identities of the CRISPR organisation. This is a precise gene editing tool that has changed how molecular biology is done, especially in terms of gene editing.
Commonly Asked Questions
Restriction enzymes are essential for molecular cloning as they allow precise cutting of DNA, enabling the insertion of genes into vectors like plasmids. This is fundamental for studying gene function and producing recombinant proteins.
Restriction enzymes allow scientists to cut specific genes from one organism and insert them into another organism's DNA. This process is crucial for creating transgenic organisms with new traits or functions.
Restriction enzymes are used to fragment an organism's entire genome into smaller pieces that can be cloned into vectors. This creates a DNA library representing all the genetic material of that organism.
Restriction enzymes allow scientists to insert genes coding for specific proteins into expression vectors. These vectors can then be introduced into bacteria or other cells to produce large quantities of the desired protein.
Restriction enzymes are crucial for creating physical maps of genomes. By cutting DNA into fragments and analyzing their sizes and order, scientists can determine the structure and organization of genomes.
Laboratory Techniques Involving Restriction Enzymes
Different techniques that involve restriction enzymes are:
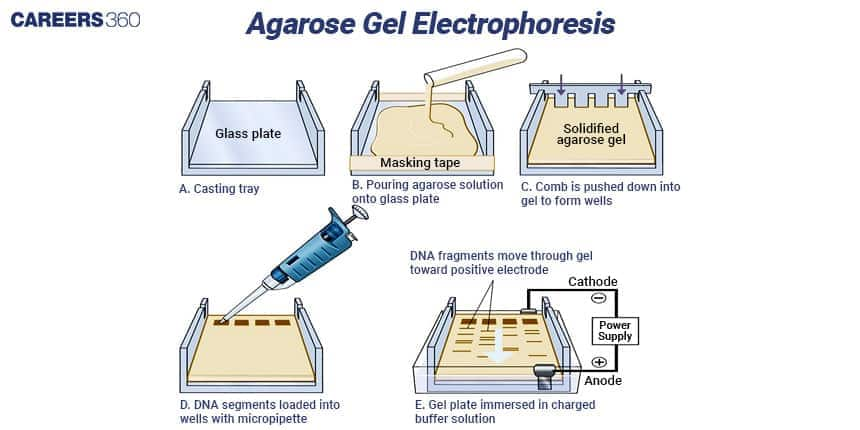
Gel Electrophoresis
The specific technique that is applied in the separation of DNA fragments is known as Gel electrophoresis. Restriction enzymes digest DNA in the presence of oxygen and thereafter the DNA fragments are applied on an agarose gel and submerged in an electric field. The DNA fragments that are smaller move in the gel much faster and can therefore be observed and quantified for fragments of choice.
DNA Ligation
A ligase catalyses the joining of DNA which is referred to as DNA ligation, where fragments of DNA are ligated to form recombinant DNA molecules. This is commonly done by using restriction enzymes that digest the DNA and DNA ligase that join compatible sticky ends. This process is very important in cloning and recombinant DNA technology.
PCR (Polymerase Chain Reaction)
PCR is a biochemical method of DNA replication that is applied in the amplification of targeted nucleic acid sequences. The desired DNA fragments of interest can be selectively amplified through the process of PCR after DNA digestion using restriction enzymes. This application is commonly employed in molecular biology, genetics, and diagnostics.
Restriction Fragment Length Polymorphism (RFLP)
RFLP analysis is a molecular method applicable to study polymorphism of nucleotide sequences in the genome. In digesting DNA with restriction enzymes the DNA samples get separated based on the size of the fragment by gel electrophoresis. In genetic mapping, RFLP analysis is broadly applied in forensic science and disease pathway diagnosis.
Commonly Asked Questions
Sticky ends are single-stranded overhangs created when some restriction enzymes cut DNA. These overhangs can easily bind to complementary sequences, making them useful in genetic engineering for joining DNA fragments.
In genetic engineering, restriction enzymes are used to cut DNA at specific sites, allowing scientists to remove, add, or replace DNA segments. This is crucial for creating recombinant DNA and genetically modified organisms.
Restriction enzymes allow scientists to cut specific genes from one organism's DNA and insert them into a plasmid or another organism's DNA. This process, called gene cloning, is fundamental to many biotechnology applications.
A restriction map is a diagram showing the locations where specific restriction enzymes cut a piece of DNA. It's useful for analyzing and manipulating DNA sequences in molecular biology.
In DNA fingerprinting, restriction enzymes cut DNA at specific sites, creating fragments of different lengths. These fragments can be separated and analyzed to create a unique "fingerprint" for an individual.
Advances and Innovations
Engineered Restriction Enzymes
Some restriction enzymes are naturally occurring, while others have been engineered to have altered specificities or even to possess different desirable attributes. These engineered enzymes add to the available repertoire of molecular/synthetic biology to seek tighter control over DNA.
Use in Synthetic Biology
In synthetic biology, restriction enzymes are indispensable since they are applied to build special DNA sequences for designing synthetic gene circuits, genetic circuits, and biosensors. The creation of synthetic DNA has various uses in biotechnology, medicine, and even environmental studies.
Development Of New Restriction Enzymes With Altered Specificity
Currently, the researchers are in the process of activity isolation and optimisation of new restriction enzymes with distinct specificities for selective DNA sequence recognition. These enzymes increase the potential of DNA manipulation techniques and facilitate the creation of new DNA constructions with desired functions.
Commonly Asked Questions
RFLPs are variations in DNA fragment lengths produced by restriction enzymes due to genetic differences between individuals. They are used in genetic mapping, disease diagnosis, and forensic science.
By cutting DNA at specific sites, restriction enzymes can reveal genetic variations between individuals or populations. This is useful in studying biodiversity, evolution, and population genetics.
In synthetic biology, restriction enzymes are used to assemble DNA parts in a modular fashion. This allows scientists to create novel genetic circuits and even synthetic genomes.
Genetic mutations can create or eliminate restriction enzyme recognition sites. By comparing restriction enzyme digestion patterns, scientists can detect these mutations, which is useful in diagnosing genetic disorders.
Restriction enzymes allow scientists to cut plasmids at specific sites and insert new genes or DNA sequences. This is crucial for creating recombinant plasmids used in various biotechnology applications.
Practical Considerations in Using Restriction Enzymes
Buffer Conditions
Some of the factors that are impacted while selecting the buffer conditions include the pH of the solution and the concentration of salts. The best buffer conditions should be chosen according to the needs of the enzyme as well as the specifics of the experiment.
Temperature Requirements
Restriction enzymes work at particular temperatures and can easily be destroyed via excessive heat. Fluctuations in temperature can impact enzymes and how effectively food is broken down. As is clear with the temperature, there are standard structures that must be followed to achieve efficient digestion of DNA.
Inhibitors and Activators
Some chemicals could influence restriction enzymes’ action; they are inhibitors or activators in this view. The effects of each of the inhibitors or activators should be closely looked at to minimise variability when using the method in DNA digestion experiments.
Troubleshooting Common Issues In Restriction Enzyme Digestion
Problems related to restriction enzymes are incomplete digestion, nonspecific digestion, and star activity. Corrective action plans could be the fine-tuning of reaction conditions, changing the enzyme concentration, or employing enzymes with other specificities.
Commonly Asked Questions
Methylases add methyl groups to specific DNA sequences in bacterial genomes. This methylation prevents restriction enzymes from cutting the bacteria's own DNA while still allowing them to cut foreign, unmethylated DNA.
Star activity refers to the reduced specificity of some restriction enzymes under non-optimal conditions, causing them to cut at sites similar but not identical to their normal recognition sequences.
In gene therapy research, restriction enzymes are used to manipulate DNA sequences, allowing scientists to insert therapeutic genes into vectors for delivery to patients or to correct faulty genes.
Blunt ends are created when restriction enzymes cut both DNA strands at exactly the same position, resulting in no overhangs. These ends are more challenging to join but can still be used in genetic engineering.
Restriction enzymes are named after the bacteria they come from. For example, EcoRI comes from Escherichia coli RY13. The first letter is from the genus, the next two from the species, and the rest identifies the strain or variant.
MCQs on Restriction Enzyme
Q1. Restriction endonucleases are enzymes which
make cuts at specific positions within the DNA molecule
recognize a specific nucleotide sequence for binding of DNA ligase
restrict the action of the enzyme DNA Polymerase
remove nucleotides from the ends of the DNA molecule
Correct answer: 1) make cuts at specific positions within the DNA molecule
Explanation:
Restriction Endonuclease - These enzymes cleave DNA only within or near the specific base sequence. These sequences are called recognition sites. They are of three types RE- I, RE-II and RE-III.
- wherein 1st Restriction enzyme (Hind II) was used in RDT.
Hence, the correct answer is option 1) make cuts at specific positions within the DNA molecule.
Q2. Which enzyme is used as a ‘molecular scissor’ in genetic engineering?
Restriction endonuclease
DNA polymerase
DNA ligase
DNA gyrase
Correct answer: 1) Restriction endonuclease
Explanation:
The enzyme used as a ‘molecular scissor’ in genetic engineering is restriction endonuclease.
Scientists can alter genes by using this enzyme to cut DNA at specified locations. It cuts DNA precisely after identifying a unique sequence. This enables researchers to modify, add, or remove genes for use in agriculture, medicine, and research. For this reason, restriction endonucleases play a crucial role in genetic engineering.
Hence, the correct answer is option 1)Restriction endonuclease.
Q3. Nuclease who removes nucleotides from the ends of the DNA in one strand of the duplex.
Exonuclease
Endonuclease
Exo-endo nuclease
None of the above
Correct answer: 1) Exonuclease
Explanation:
Exonucleases are enzymes that specifically remove nucleotides from the ends of DNA strands. They function by cleaving nucleotides one at a time from either the 5′ end or the 3′ end of a single strand within a DNA duplex. This precise activity makes exonucleases important for various cellular processes, including DNA repair, replication, and degradation of unneeded or damaged DNA.
Hence the correct answer is option 1) Exonuclease.
Also Read:
Frequently Asked Questions (FAQs)
Q1. What are restriction enzymes?
Restriction enzymes are molecular scissors that cut DNA at specific recognition sequences. They are widely used in genetic engineering and recombinant DNA technology.
Q2. What are Type 1 and Type 2 restriction enzymes?
Type 1 enzymes cut DNA at random sites far from their recognition site, while Type 2 enzymes cut at specific sites within or close to their recognition sequence. Type 2 enzymes are more commonly used in biotechnology.
Q3. Why are two restriction enzymes used?
Using two different restriction enzymes allows precise cutting at two distinct sites, which helps in directional cloning and prevents the vector from re-ligating without the insert.
Q4. What is the difference between blunt and sticky ends?
Blunt ends are straight cuts with no overhangs, while sticky ends have short single-stranded overhangs. Sticky ends are easier to join during cloning due to base pairing.
Q5. What is the full form of ECOR1?
ECOR1 stands for Escherichia coli RY13 Restriction enzyme 1, where "Eco" refers to the bacterial species and "R" denotes the strain.
Recommended Video on Restriction Endonuclease
Frequently Asked Questions (FAQs)
By comparing restriction enzyme digestion patterns across species, scientists can infer genomic rearrangements and other evolutionary changes, providing insights into genome evolution over time.
Restriction enzymes are crucial for engineering viral and non-viral vectors used in gene therapy. They allow scientists to insert therapeutic genes into these vectors for delivery to target cells.
Restriction enzymes can be used to analyze the DNA of different bacterial species, helping scientists identify genes that have been transferred horizontally between unrelated organisms.
In DNA nanotechnology, restriction enzymes are used to prepare DNA strands for creating complex 3D structures known as DNA origami, which have potential applications in drug delivery and molecular computing.
Restriction enzymes can create specific DNA damage, allowing scientists to study how cells repair different types of DNA lesions and understand DNA repair pathways.
Restriction enzymes are used in the development of DNA-based biosensors. They help in creating specific DNA sequences that can detect target molecules or environmental conditions.
Restriction enzymes can be used in techniques like Southern blotting to detect chromosomal abnormalities such as deletions, duplications, or translocations.
Restriction enzymes allow scientists to modify genes coding for proteins. This enables the creation of fusion proteins, truncated proteins, or proteins with altered properties for various applications.
Restriction enzymes are used to modify DNA sequences for creating knockout organisms, where specific genes are made inoperative. This helps scientists study gene function.
In metagenomics, restriction enzymes are used to fragment environmental DNA samples. This allows for the creation of metagenomic libraries and the study of microbial communities in various environments.